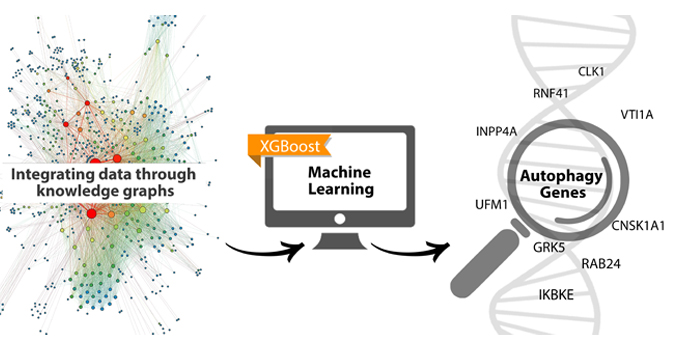
In a groundbreaking study conducted by University of New Mexico researchers, scientists have harnessed the power of machine learning to identify a set of previously unknown genes associated with autophagy, a vital cellular process involved in recycling and maintaining cellular health.
Leveraging a state-of-the-art machine learning model, the study identified 193 genes as potential contributors to autophagy machinery. These previously overlooked “dark genes” represent promising avenues for unraveling the mysteries of autophagy and its role in cellular functioning and complex diseases such as Alzheimer's, said UNM neuroscientist Elaine Bearer, MD, PhD.
“This is another form of unbiased, data-driven science,” Bearer said. “What machine learning is allowing us to do is avoid the guesswork and do discovery science in a non-hypothesis-driven way.”
The study, entitled "Autophagy Dark Genes: Can We Find Them with Machine Learning?" was recently published in the journal Natural Sciences, and aimed to identify an autophagy-related gene set by combining diverse biological features and datasets and plugging the data into an artificial intelligence algorithm.
“The idea was, ‘Can we find these dark, hidden, secret genes with an artificial intelligence investigation?’” Bearer said.
The answer is yes, machine learning can guide genomics research to gain a more complete annotation of complex processes.
But machine learning is not the end of the task, Bearer emphasizes. Once the artificial intelligence has identified something, it’s up to scientists to validate both the process and the results.
To accomplish this, a research team at UNM employed the MetaPath/XGBoost (MPxgb) machine learning model, which was trained using data from 17 different sources. The artificial intelligence research investigation began in 2019, led by Tudor Oprea, MD, PhD, former director for Screening Informatics for UNM’s Center for Molecular Discovery and Drug Discovery Core and a member at the UNM Comprehensive Cancer Center.
Mohsen Ranjbar, PharmD, a UNM graduate student in chemistry and chemical biology, took Oprea’s research and conducted a validation search, combing through the Autophagy Database and through research publication databases, like PubMed, to see if the model demonstrated high accuracy in distinguishing already-known autophagy-associated genes.
We can use machine learning more than before. Sometimes we have a limited knowledge on something, but we can use machine learning to shed light on things and give us directions going forward.
Through the search, Ranjbar’s findings revealed that while 23% of the top predicted genes were already annotated in the Autophagy Database, a staggering 77% (193 genes) were novel discoveries, representing an untapped potential for understanding autophagy regulation in cellular processes.
“It’s interesting and it was surprising,” Ranjbar said. “It’s only been a short time since we began this research, and to see that some of these specific AI-discovered genes have already been mentioned as newly discovered autophagy genes in different recent publications, it shows the validation of our machinery to find these genes.”
Bearer said that by uncovering these autophagy dark genes, researchers can delve deeper into the relationship between autophagy dysregulation and the development of diseases, ultimately guiding the development of new therapeutic strategies for the disease.
The groundbreaking study also showcases the versatility of machine learning and artificial intelligence in genomic research, extending knowledge beyond autophagy into other areas of biology.
“We don’t know all the genes involved in things like endosomal trafficking, which is really important in lots of diseases, including Alzheimer’s disease,” Bearer said. “So, we could use our machine learning model to investigate and identify other genes in the genome that have not yet had a wet-bench test for what their functional role is.”
The study was made possible through the support from several grants, including NIH U24CA224370, U24TR002278, UL1TR001449, P20GM121176, P20AG068077, R01 MD014153 and the Harvey Family Endowment.
Additional support was provided from the New Mexico Alzheimer’s Disease Research Center, the UNM Autophagy, Inflammation & Metabolism Center, and the UNM Clinical & Translational Science Center.
Bearer said the interdisciplinary study wouldn’t have been possible without having crossed academic and research department boundaries. She works in the Department of Pathology, Ranjbar is in the Chemistry department, and other contributors to the project were in Internal Medicine, Computer Science and the Molecular Discovery Center.
“This big project transcended multiple entities within UNM,” she said. “I want to influence the scientific thinking around the use of machine learning, because it’s so powerful.”